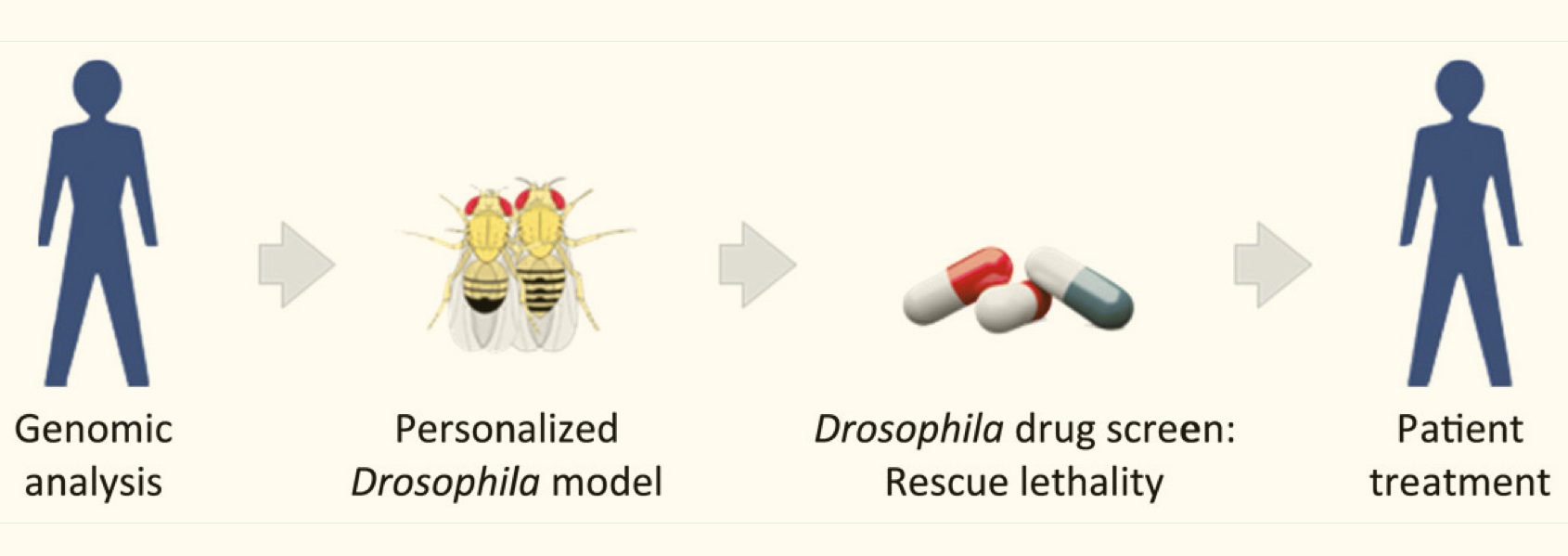
Diversity in Clinical Genetics Remains Poorly Defined
/Diversity in Clinical Genetics Remains Poorly Defined but the Clinical Genome Resource (ClinGen) Ancestry and Diversity Working Group is working to address this important issue. In clinical genetics and genomics, many approaches depend on the ability to identify genetic variation that appears to be non-randomly distributed in a population. However, genetic variation often clusters in ways that reflect how peoples’ ancestors were grouped together. These historical associations are often summarized by the terms Race, Ethnicity, and Ancestry but what these terms mean, both semantically and biologically, are still very unclear. The paper below provides no clear solutions but is an excellent introduction and discussion of this problem and the challenges we face in addressing it.
Clinical Genetics Lacks Standard Definitions and Protocols for the Collection and Use of Diversity Measures
Abstract
Genetics researchers and clinical professionals rely on diversity measures such as race, ethnicity, and ancestry (REA) to stratify study participants and patients for a variety of applications in research and precision medicine. However, there are no comprehensive, widely accepted standards or guidelines for collecting and using such data in clinical genetics practice. Two NIH-funded research consortia, the Clinical Genome Resource (ClinGen) and Clinical Sequencing Evidence-generating Research (CSER), have partnered to address this issue and report how REA are currently collected, conceptualized, and used. Surveying clinical genetics professionals and researchers (n = 448), we found heterogeneity in the way REA are perceived, defined, and measured, with variation in the perceived importance of REA in both clinical and research settings. The majority of respondents (>55%) felt that REA are at least somewhat important for clinical variant interpretation, ordering genetic tests, and communicating results to patients. However, there was no consensus on the relevance of REA, including how each of these measures should be used in different scenarios and what information they can convey in the context of human genetics. A lack of common definitions and applications of REA across the precision medicine pipeline may contribute to inconsistencies in data collection, missing or inaccurate classifications, and misleading or inconclusive results. Thus, our findings support the need for standardization and harmonization of REA data collection and use in clinical genetics and precision health research.