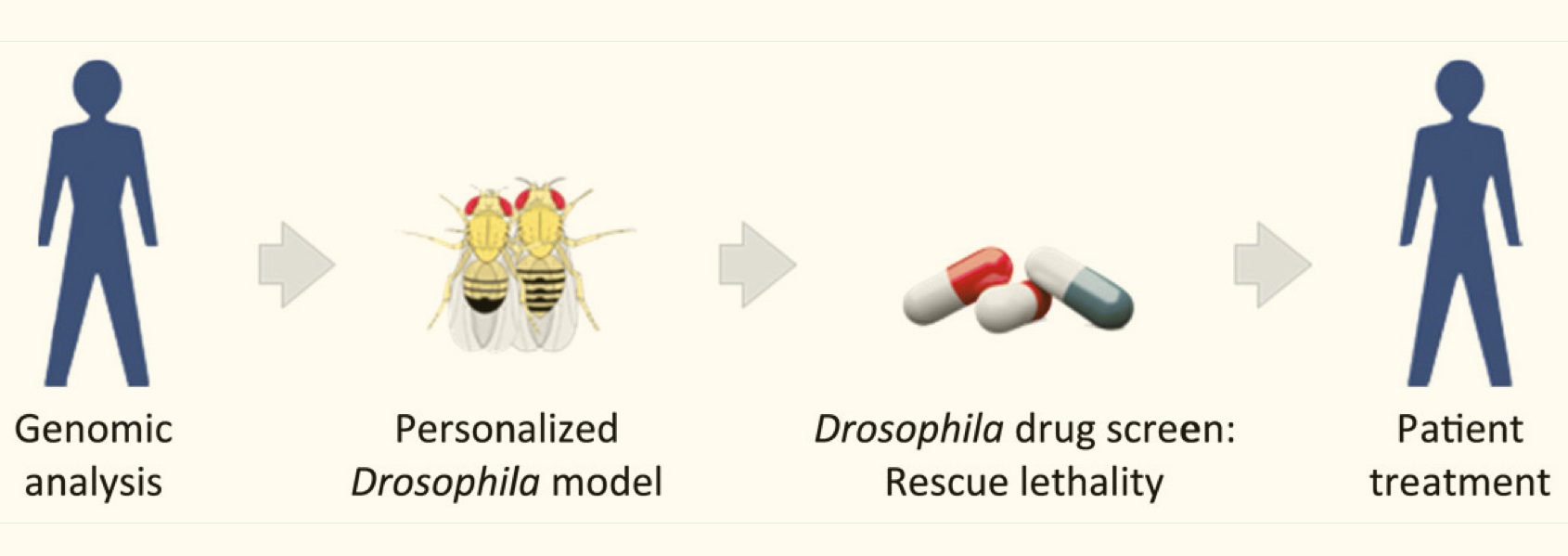
A Primer on Cancer from the NIH National Cancer Institute
/Here is an excellent resource for understanding and explaining cancer, directly from the National Institute of Health National Cancer Institute.
A dividing breast cancer cell. Credit: National Cancer Institute / Univ. of Pittsburgh Cancer Institute
What is Cancer? A Collection of Related Diseases
Cancer is the name given to a collection of related diseases. In all types of cancer, some of the body’s cells begin to divide without stopping and spread into surrounding tissues.Cancer can start almost anywhere in the human body, which is made up of trillions of cells. Normally, human cells grow and divide to form new cells as the body needs them. When cells grow old or become damaged, they die, and new cells take their place.
When cancer develops, however, this orderly process breaks down. As cells become more and more abnormal, old or damaged cells survive when they should die, and new cells form when they are not needed. These extra cells can divide without stopping and may form growths called tumors.
Many cancers form solid tumors, which are masses of tissue. Cancers of the blood, such as leukemias, generally do not form solid tumors.
Cancerous tumors are malignant, which means they can spread into, or invade, nearby tissues. In addition, as these tumors grow, some cancer cells can break off and travel to distant places in the body through the blood or the lymph system and form new tumors far from the original tumor.
Unlike malignant tumors, benign tumors do not spread into, or invade, nearby tissues. Benign tumors can sometimes be quite large, however. When removed, they usually don’t grow back, whereas malignant tumors sometimes do. Unlike most benign tumors elsewhere in the body, benign brain tumors can be life threatening.
Differences between Cancer Cells and Normal Cells
Cancer cells differ from normal cells in many ways that allow them to grow out of control and become invasive. One important difference is that cancer cells are less specialized than normal cells. That is, whereas normal cells mature into very distinct cell types with specific functions, cancer cells do not. This is one reason that, unlike normal cells, cancer cells continue to divide without stopping.