Clinical application of tumor evolution analysis
/Translating insights into tumor evolution to clinical practice: promises and challenges
Matthew W. Fittall and Peter Van Loo
Genome Medicine (Review Article)
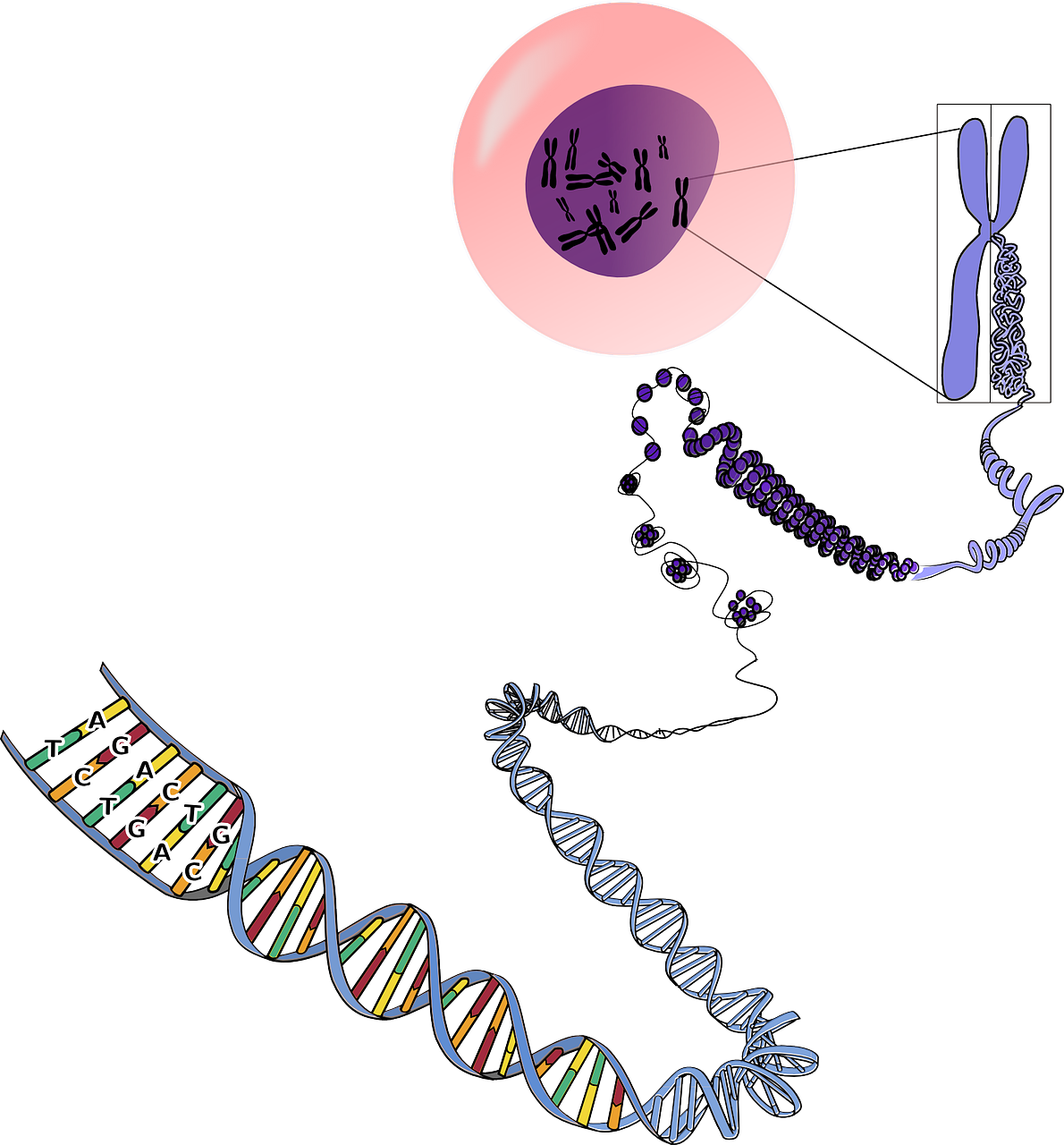
Abstract—Accelerating technological advances have allowed the widespread genomic profiling of tumors. As yet, however, the vast catalogues of mutations that have been identified have made only a modest impact on clinical medicine. Massively parallel sequencing has informed our understanding of the genetic evolution and heterogeneity of cancers, allowing us to place these mutational catalogues into a meaningful context. Here, we review the methods used to measure tumor evolution and heterogeneity, and the potential and challenges for translating the insights gained to achieve clinical impact for cancer therapy, monitoring, early detection, risk stratification, and prevention. We discuss how tumor evolution can guide cancer therapy by targeting clonal and subclonal mutations both individually and in combination…
READ MORE …