Cloud-based access to the fully sequenced genomes of 10,000 pediatric patients with cancer
/Genomics Data Could Lead to New Treatments for Children
BY BETH FAND INCOLLINGO
PUBLISHED MARCH 12, 2019
St. Jude Children’s Research Hospital is offering cloud-based access to the fully sequenced genomes of 10,000 pediatric patients with cancer, in the hopes that sharing the information will lead to the highest possible number of treatment breakthroughs.
Called the Pediatric Cancer Genome Project (PCGP), the growing set of data, categorized by cancer type, is meant to help researchers at the Memphis facility and beyond understand the genetic mutations that drive pediatric cancers and find new drugs to treat the diseases.
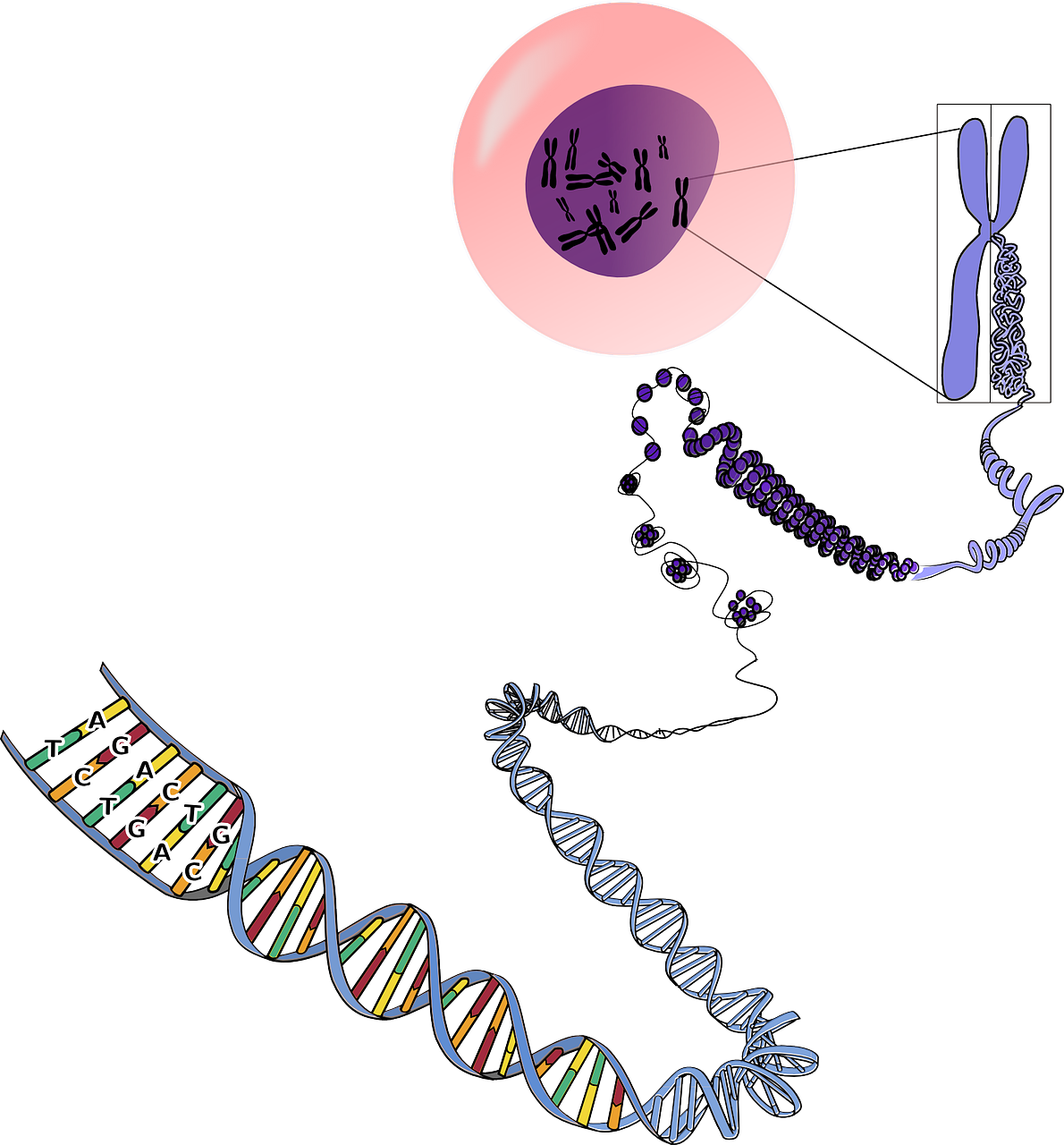
In whole-genome sequencing, a child’s normal and tumor genes are sequenced and then compared. Mutations that are present in a child’s tumor but not his or her normal genes may be driving the disease, and could be good candidates to target with drugs, said Jinghui Zang, Ph.D., chair of the Department of Computational Biology at St. Jude.