Categorizing Cells with Machine Learning and Latent Space
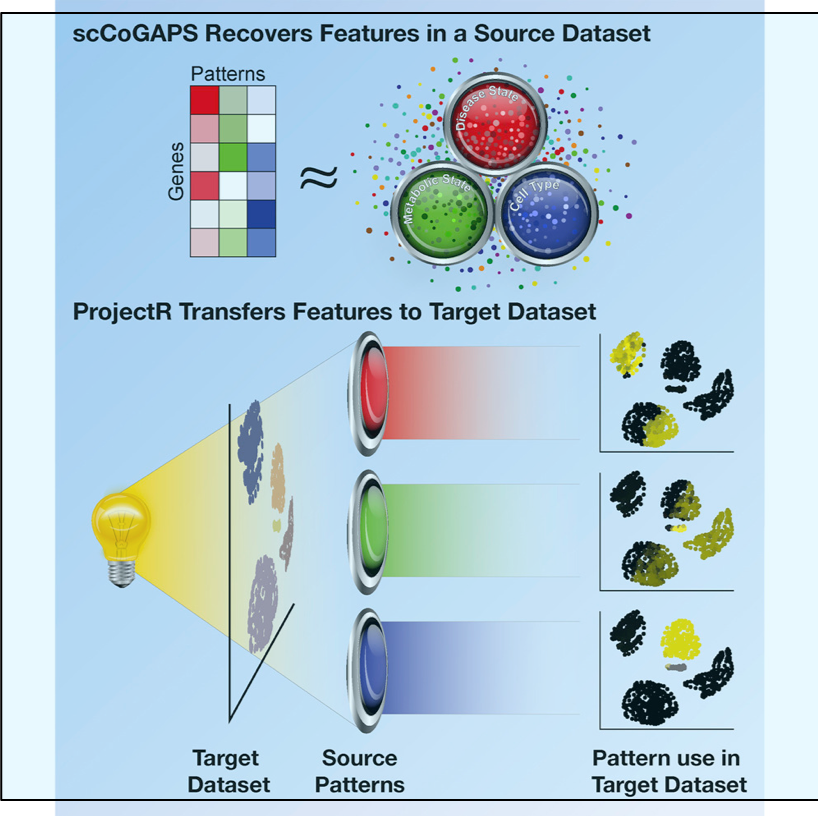
/Two exciting and complementary machine learning methods for assigning cell identity based on single-cell sequencing data were published in a paper from Johns Hopkins. The first program, scCoGAPS, defines latent spaces from a single-cell RNA-sequencing dataset to categorize cells and the second program, projectR, evaluates latent spaces in independent target datasets using transfer learning. These two methods are interesting advances towards a goal that is likely still far off—understanding exactly what makes each cell what it is. For an excellent summary read the press release, Finding A Cell’s True Identity.
The original article is a more complicated reading but interesting through out.
Stein-O’Brien, et al. (2019) Decomposing Cell Identity for Transfer Learning across Cellular Measurements, Platforms, Tissues, and Species. Cell Systems
Summary
Analysis of gene expression in single cells allows for decomposition of cellular states as low-dimensional latent spaces. However, the interpretation and validation of these spaces remains a challenge. Here, we present scCoGAPS, which defines latent spaces from a source single-cell RNA-sequencing (scRNA-seq) dataset, and projectR, which evaluates these latent spaces in independent target datasets via transfer learning. Application of developing mouse retina to scRNA-Seq reveals intrinsic relationships across biological contexts and assays while avoiding batch effects and other technical features. We compare the dimensions learned in this source dataset to adult mouse retina, a time-course of human retinal development, select scRNA-seq datasets from developing brain, chromatin accessibility data, and a murine-cell type atlas to identify shared biological features. These tools lay the groundwork for exploratory analysis of scRNA-seq data via latent space representations, enabling a shift in how we compare and identify cells beyond reliance on marker genes or ensemble molecular identity.